Engineering enzymes to improve biofuels production, and watching enzymes evolve is all in a day’s work for University of Otago protein biochemist Wayne Patrick.
He says that what he observes in the laboratory “is a good mimic of the evolution that’s happening around us all the time.”
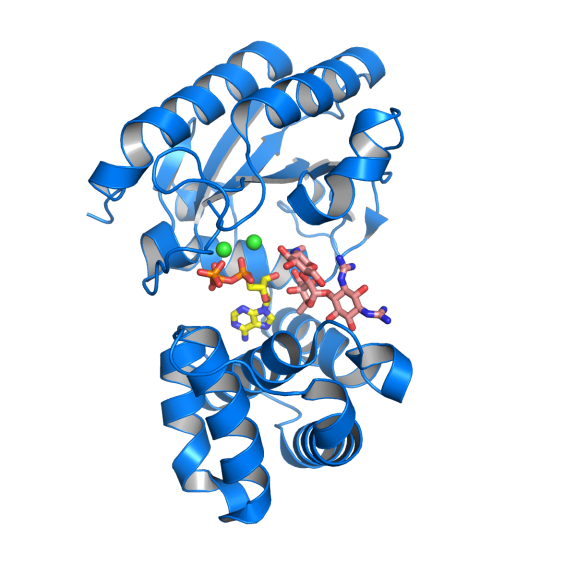
Antibiotic resistance can develop in a number of ways. In this example, the antibiotic resistance enzyme AadA (blue ribbon), targets the drug streptomycin (salmon colour). It inactivates the drug by linking an ATP molecule (yellow) to it. As a result the “enlarged” drug no longer fits in its binding site in the drug target, the ribosome. Two important magnesium ions are in green. Photo: Maria Selmer / Uppsala University
The big advantage is that it is evolution that happens very quickly - it is possible to watch proteins change over thousands of generations in just a few weeks or months. And it is also possible to see the structural impact of evolutionary changes at an atomic level.
Understanding how evolution works at this scale is relevant to understanding how microbes such as bacteria are able to rapidly evolve resistance to antibiotics.
Many familiar groups of antibiotics, such as tetracyclines and streptomycin, disrupt the ability of bacteria to produce the proteins that are vital to their survival.
Maria Selmer is a protein chemist from the University of Uppsala in Sweden, and she says that the ribosome in both human and bacterial cells play a key role in producing proteins. Small differences between the structure of bacterial and human ribosomes, however, mean that an antibiotic can lock on and disable a bacterial ribosome without affecting human cells.
“For a biochemist the ribosome is an enormous and complicated molecular machine,” says Patrick. ”It’s much bigger than the other enzymes that we work on, whereas antibiotics are relatively tiny, just a small number of atoms.”
Selmer says that the first atomic pictures of the ribosome were created in the early 2000s, and the hope was that it would now be a simple matter to predict good places for antibiotics to dock onto, leading perhaps to a boom in the development of new antibiotics. However, she says it hasn’t been that easy as the ribosome is too large and complex.
“You can make the analogy of a large lecture theatre filled with a thousand seats, and this tiny molecule (the antibiotic) will just fit in one of them – but it’s really tricky to predict which one.”
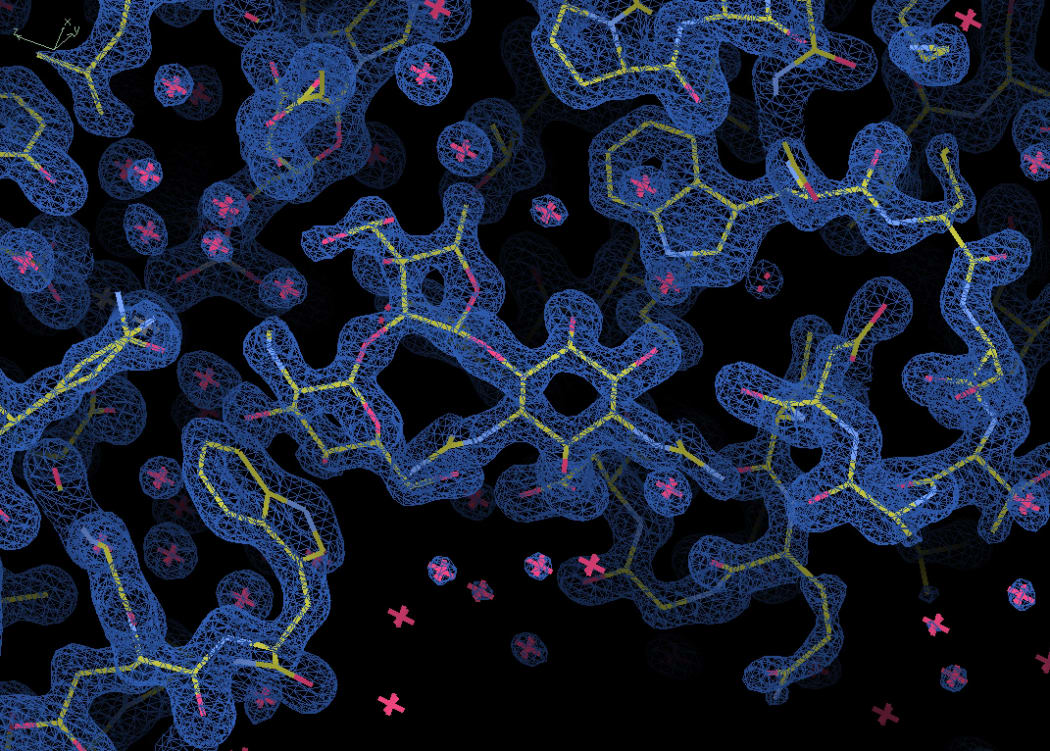
An electron density map of part of the antibiotic resistance enzyme AadA. The central shape is a molecule of the antibiotic streptomycin, which is designed to lock onto this particular binding site. Photo: Maria Selmer / Uppsala University
Patrick and Selmer say that antibiotic resistance develops in many different ways.
Selmer has been working on an antibiotic resistance enzyme called AadA, which targets the drug streptomycin. It inactivates the drug by linking an ATP molecule to it. As a result the “enlarged” drug no longer fits in its binding site on the ribosome. Building on her analogy of an antibiotic fitting just one seat in a large lecture theatre, Selmer says it's as if the antibiotic has been made to wear a thick coat so it no longer fits the seat.
They have been collaborating on a project that might enable two existing antibiotics to be used together in a way that overcomes bacterial resistance. They also believe that the ability to look at protein structure at an atomic level will help in the creation of new antibiotics in the future.
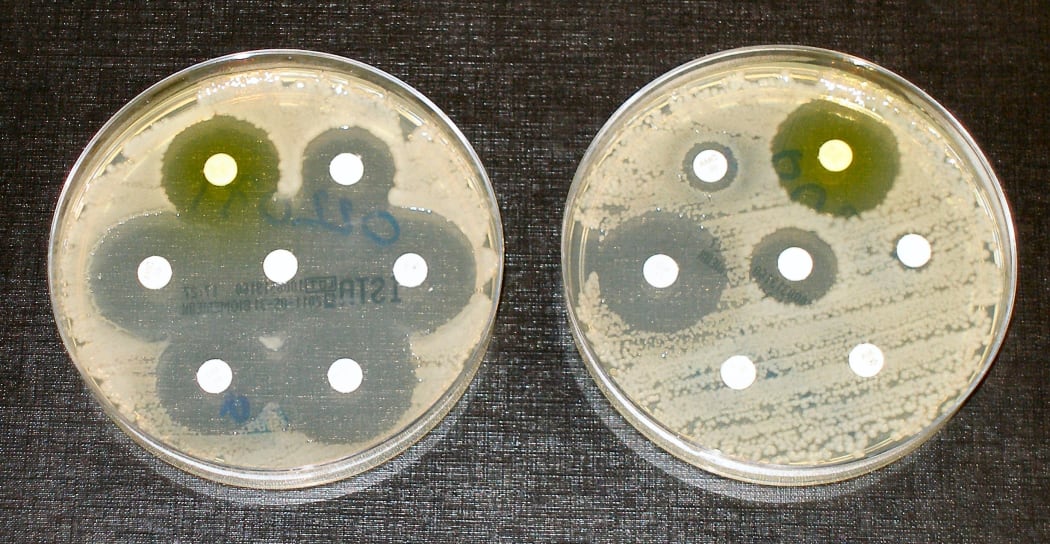
To measure antibiotic resistance bacteria are streaked on dishes with white antibiotic impregnated disks. Clear rings, such as those on the left, show that bacteria have not grown—indicating that the bacteria are not resistant to antibiotics. The bacteria in the petri dish on the right fully susceptible to only three of the seven antibiotics tested. Photo: CC BY-SA 4.0 Graham Beards
The issue of antibiotic resistance is in the news again this week, with the release by the Royal Society Te Apārangi of a special report: Antimicrobial resistance – implications for New Zealand, which stresses that it is a global health problem that will require significant international cooperation to solve.
University of Auckland microbiologist Siouxsie Wiles has recently published a book called Antibiotic resistance - the end of modern medicine? (Bridget William Books 2017).